Boost Spatial Data Science Workflows with GRASS GIS and R
Veronica Andreo and the GRASS GIS community
NCSU GeoForAll Lab
at the
Center for Geospatial Analytics
NC State University
useR! Conference, Salzburg, July 8-11, 2024

After a talk with Jakub Nowosad at FOSS4G Europe last week :)
About the presenter
- Visiting scholar at NC State's Center for Geospatial Analytics
- Lic. & PhD in Biology, MSc in Applications of the Spatial Information
- Researcher & lecturer at Gulich Institute in Argentina
- GRASS GIS: Development Team, PSC chair
- OSGeo: Charter member

What is GRASS GIS?
What is GRASS GIS?
Geek open-source command line GIS
What is GRASS GIS?
Open-source desktop GIS
What is GRASS GIS?
Processing backend in QGIS
 Image source: baharmon.github.io/grass-in-qgis
Image source: baharmon.github.io/grass-in-qgis
What is GRASS GIS?
Data analytics tool in R or quarto notebooks
What is GRASS GIS?
Geovisualization and data analytics tool in Python notebooks
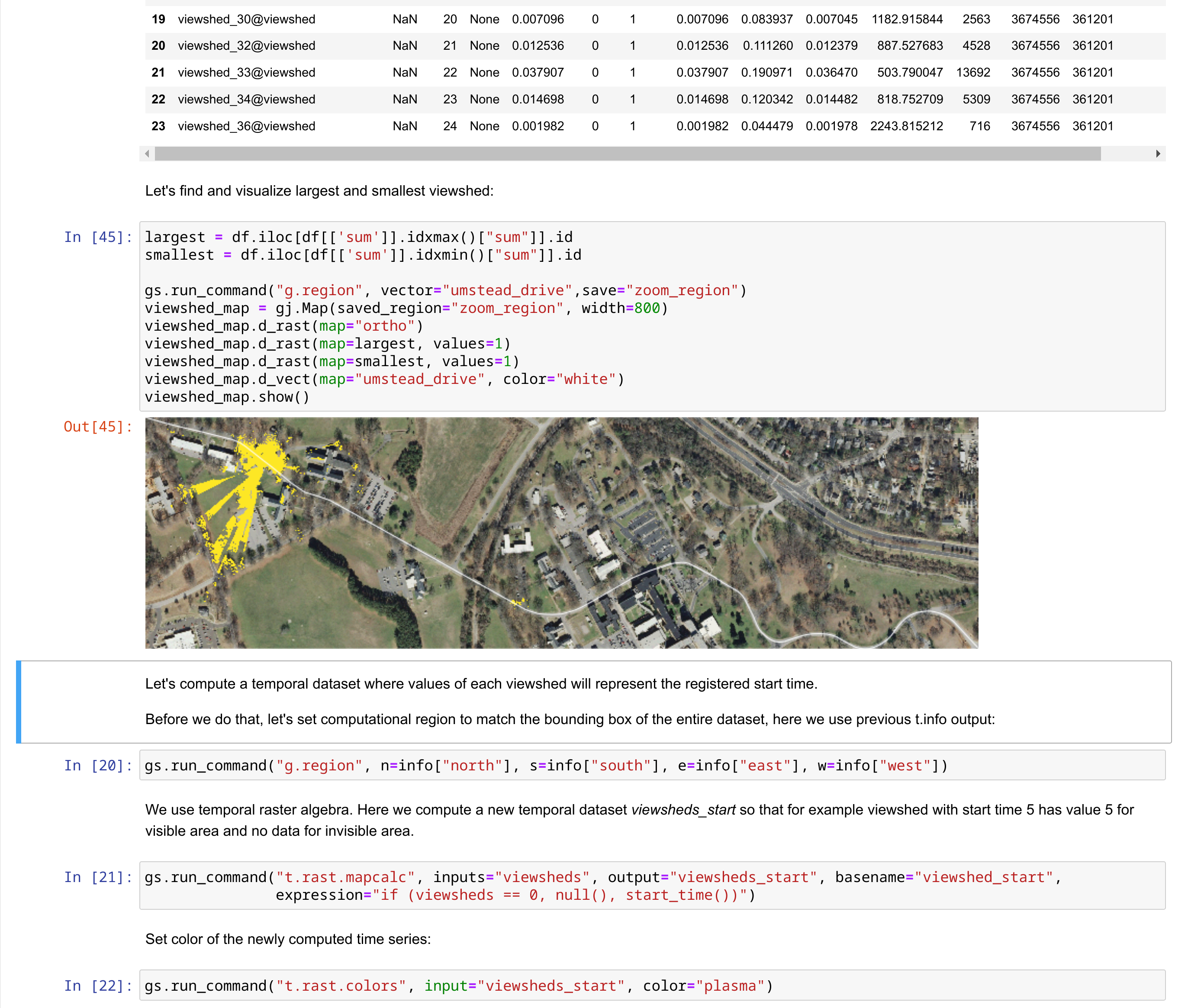
What is GRASS GIS?
Geoprocessing engine running in an HPC environment
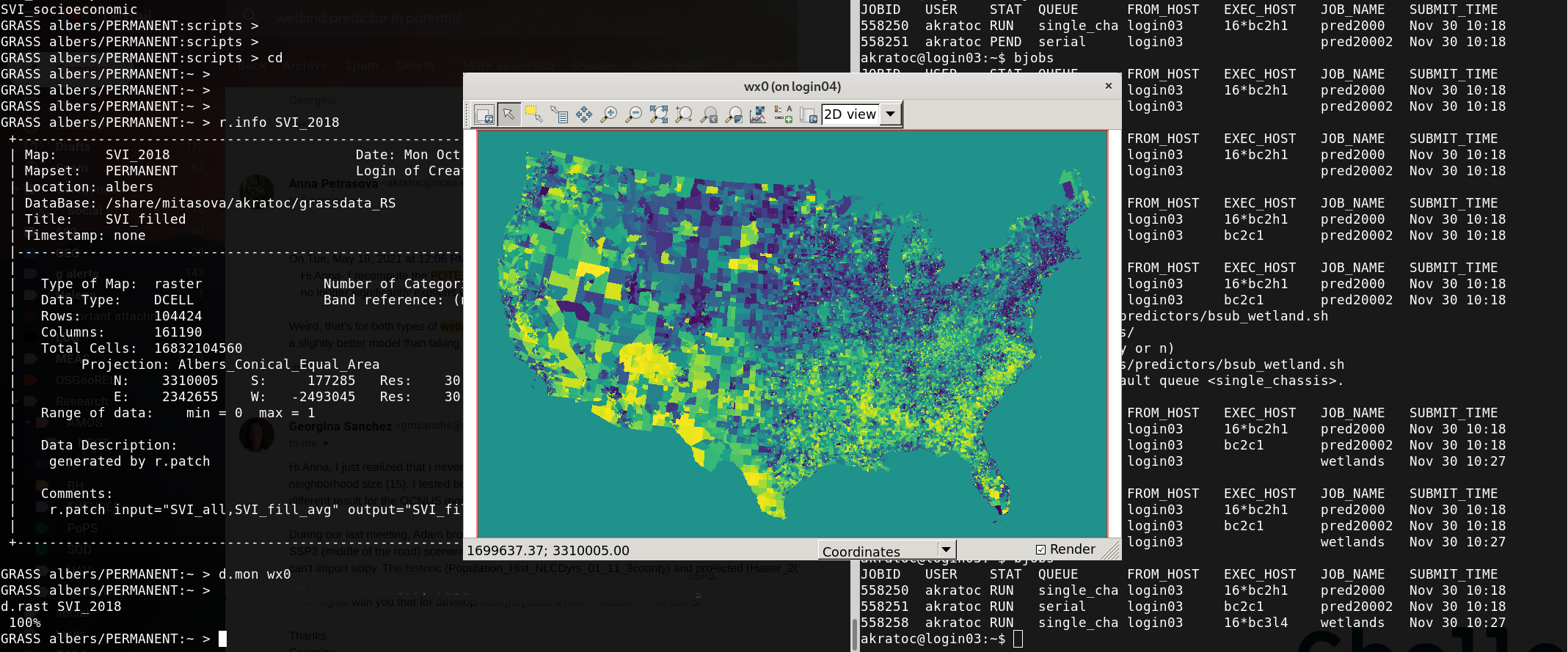
What is GRASS GIS?
Geospatial platform for developing custom models
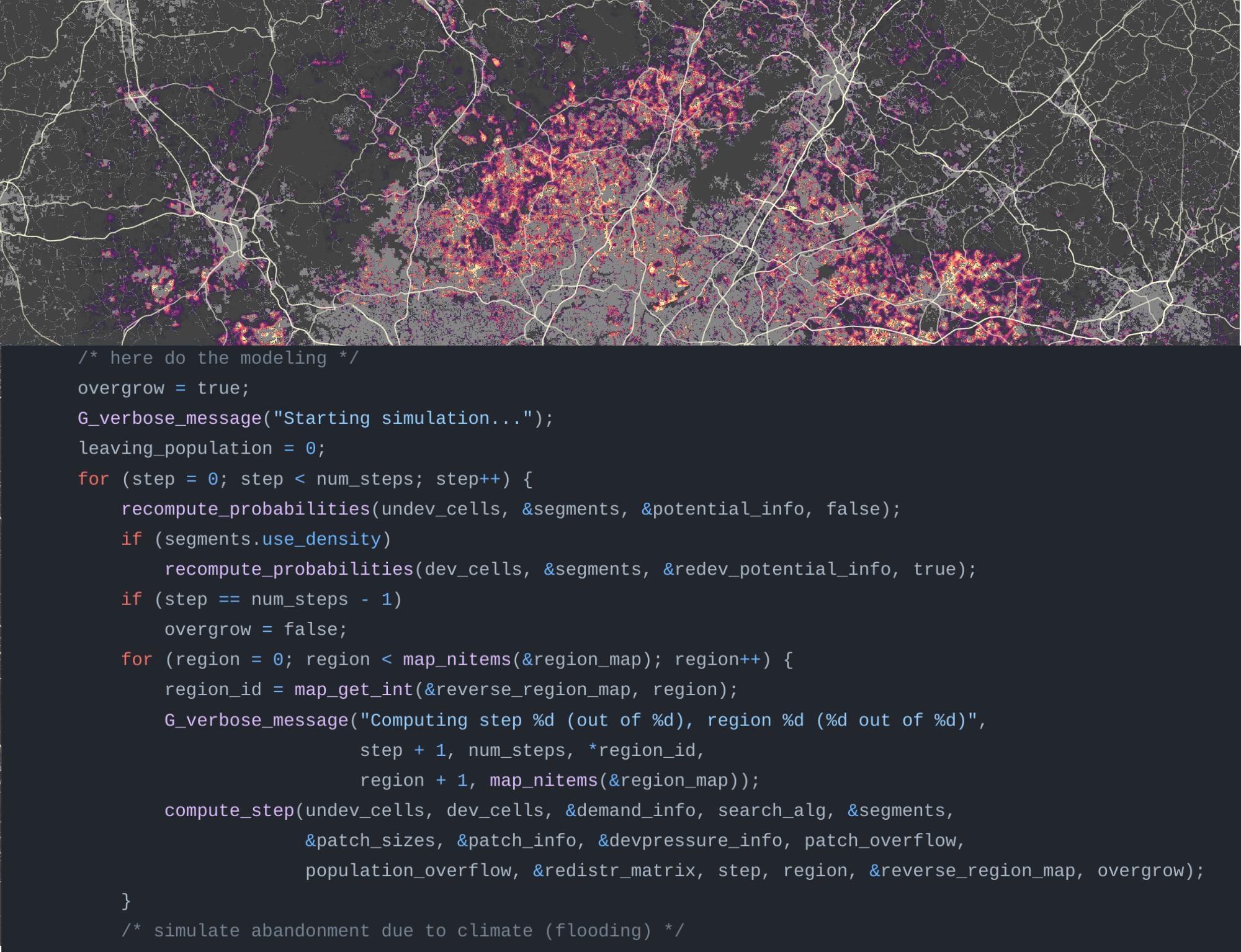
What is GRASS GIS?
Cloud geoprocessing backend
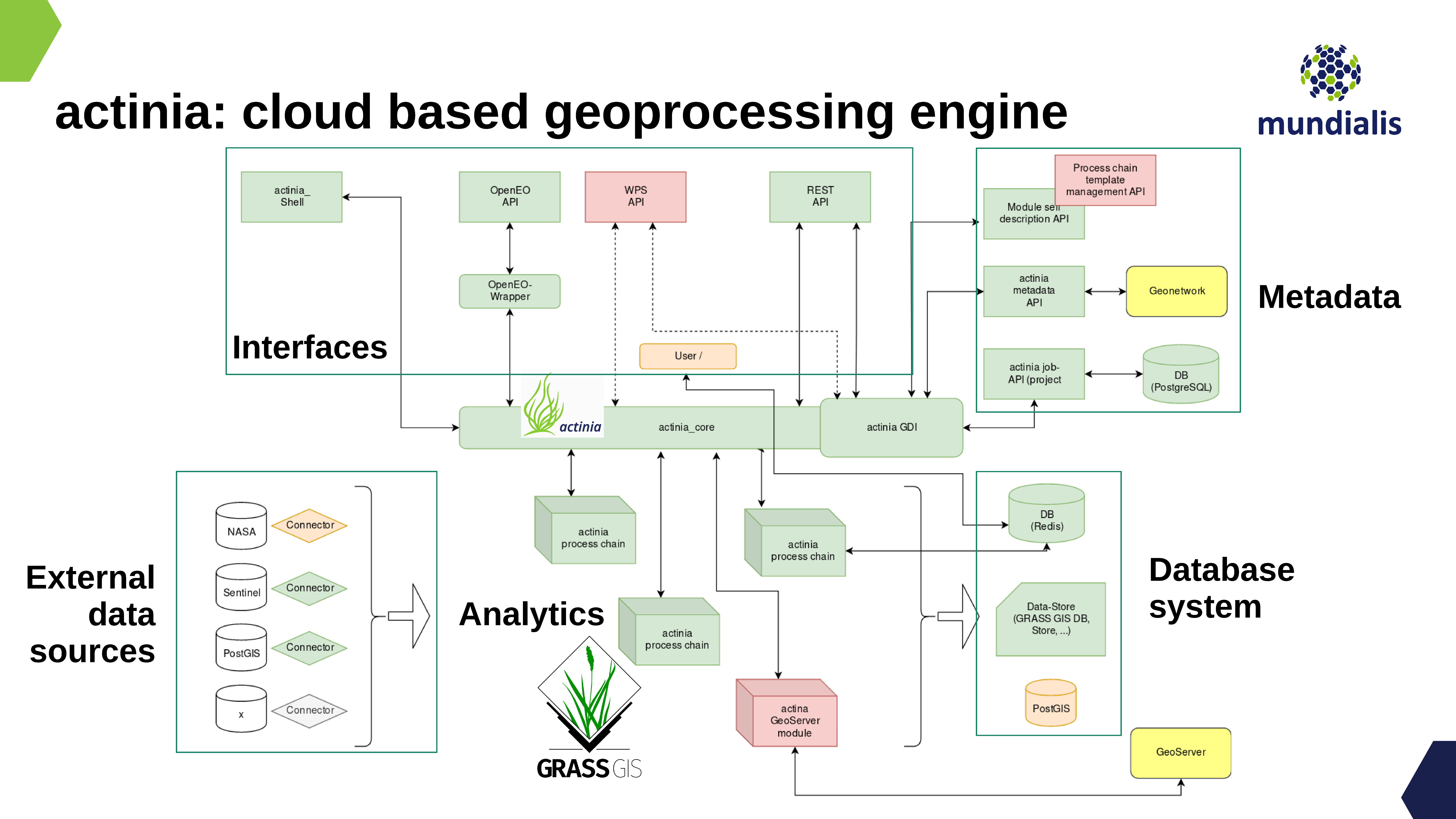 Image source: neteler.gitlab.io/actinia-introduction/
Image source: neteler.gitlab.io/actinia-introduction/
GRASS is all of that!
Robustness, Innovation with Stability, Versatility, Scientific foundations
Why using GRASS?
Mature and robust geoprocessing engine
- 500+ tools in core, 400+ addons
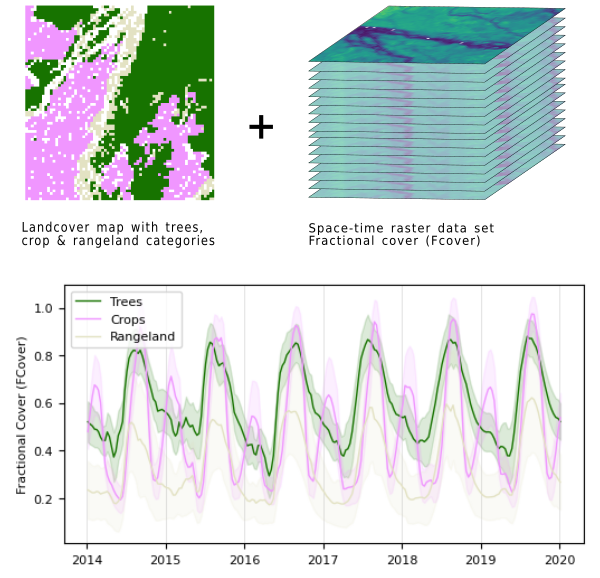

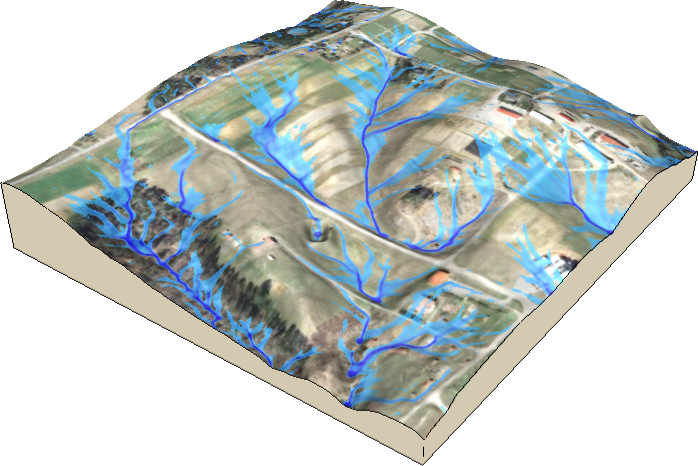
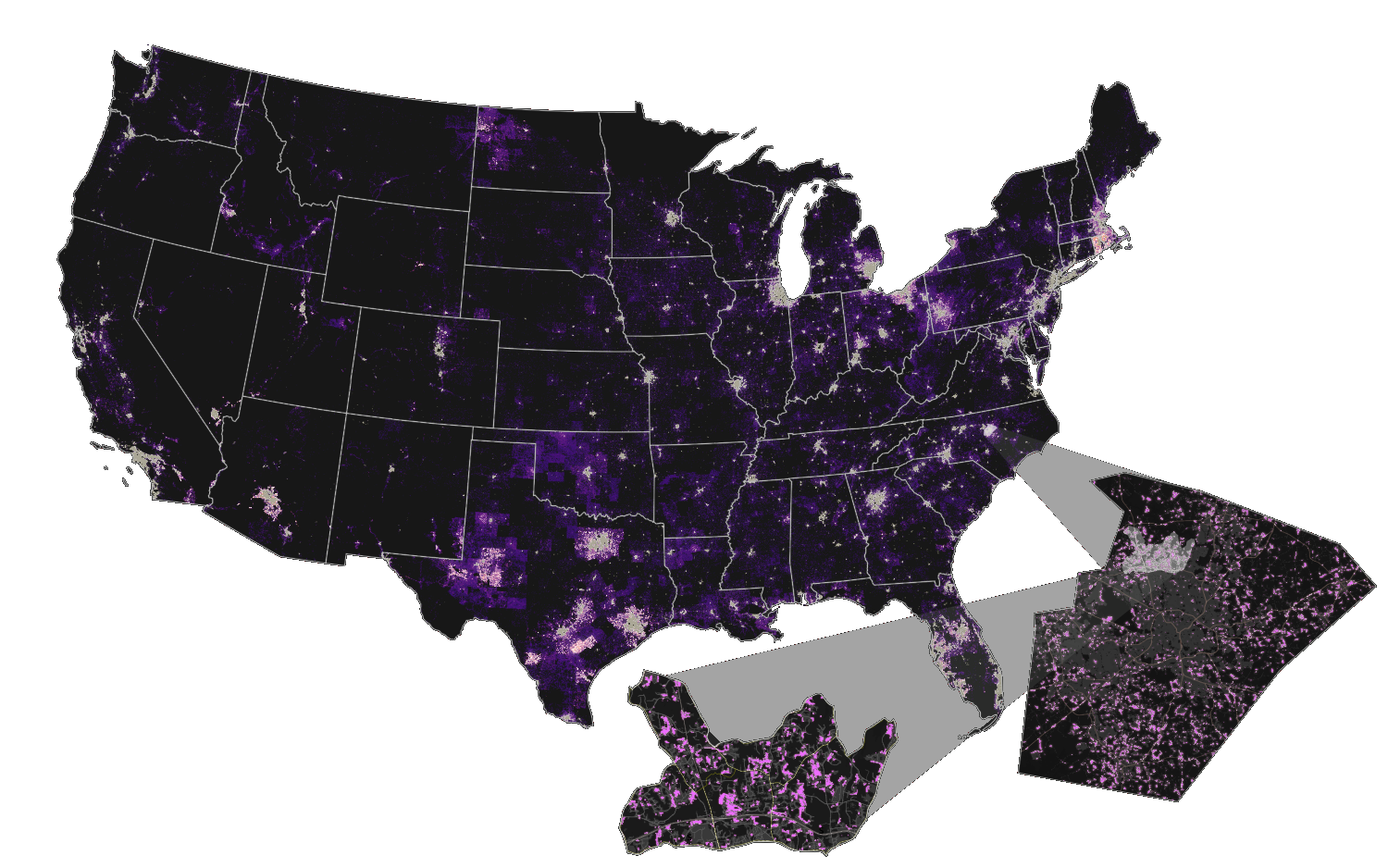
Unique tools for diverse applications!!
Tools with parallel support
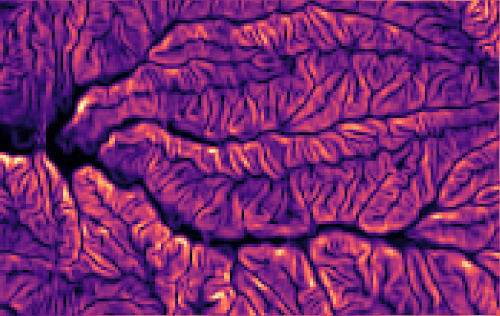 r.slope.aspect
r.slope.aspect
 r.neighbors
r.neighbors
 r.mfilter
r.mfilter
 r.series
r.series
 r.patch
r.patch
 r.sun
r.sun
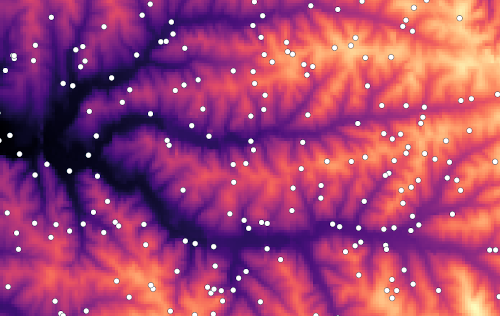 v.surf.rst
v.surf.rst
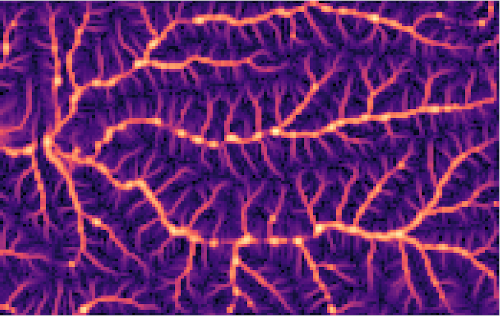 r.sim.water
r.sim.water
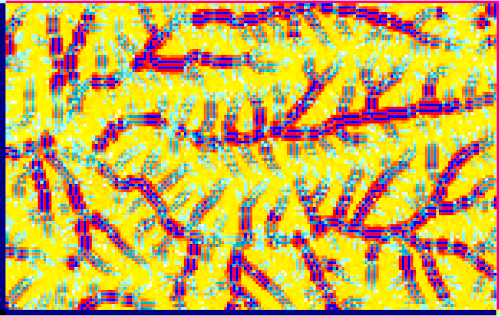 r.sim.sediment
r.sim.sediment
 r.resamp.interp
r.resamp.interp
 r.resamp.filter
r.resamp.filter
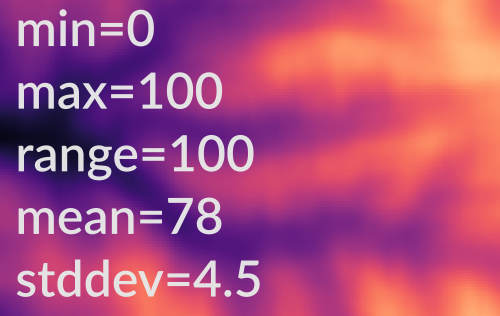 r.univar
r.univar
And more at grass.osgeo.org/grass-stable/manuals/keywords.html#parallel
API tooling for parallelizing Python scripts
- Data parallelization: GridModule
- Task parallelization: multiprocessing, ParallelModuleQueue
from grass.pygrass.modules.grid import GridModule
grd = GridModule("v.to.rast", input="roads", output="roads",
use="val", processes=4)
grd.run()
r.mapcalc.tiled "log_dist = if (dist == 0, 0, log(dist))" nprocs=4
HPC ready
- Recipes for High Performance Computing setups
- Better integration with conda environments: conda package on the way!
- Reduced potential for race conditions
- Easily create parallel workflows
 by Vaclav Petras, Markus Neteler, Nicklas Larsson, …
by Vaclav Petras, Markus Neteler, Nicklas Larsson, …
Streamlined access to data
Automated download and import of diverse datasets i.eodag, i.sentinel, i.modis, i.landsat, r.in.usgs, r.in.nasadem, m.crawl.thredds, t.rast.import.netcdf, t.stac (on the way!), …
by Fondazione Edmund Mach, OpenGeoLabs, mundialis, GSoC, NC State University, CONICET, Norwegian Institute for Nature Research, …
Optimized for large data
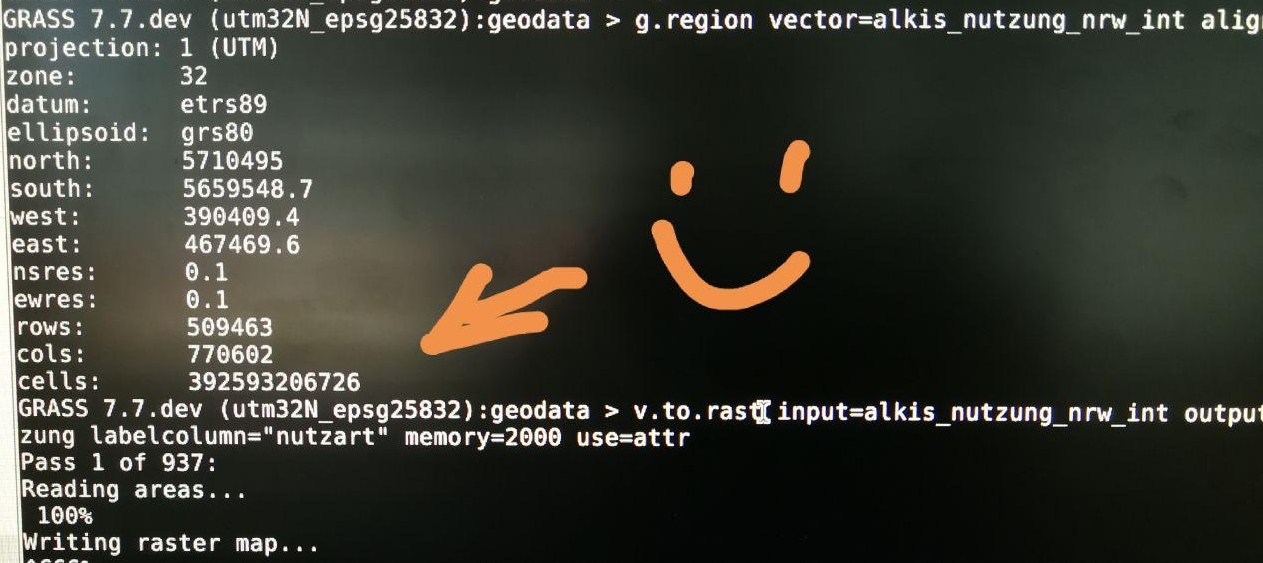
392 billion cells!
Fast External Data Links
r.external links (opens) external raster data (GeoTiffs, …) much faster.(2-5× faster, or almost no time for some workflows)
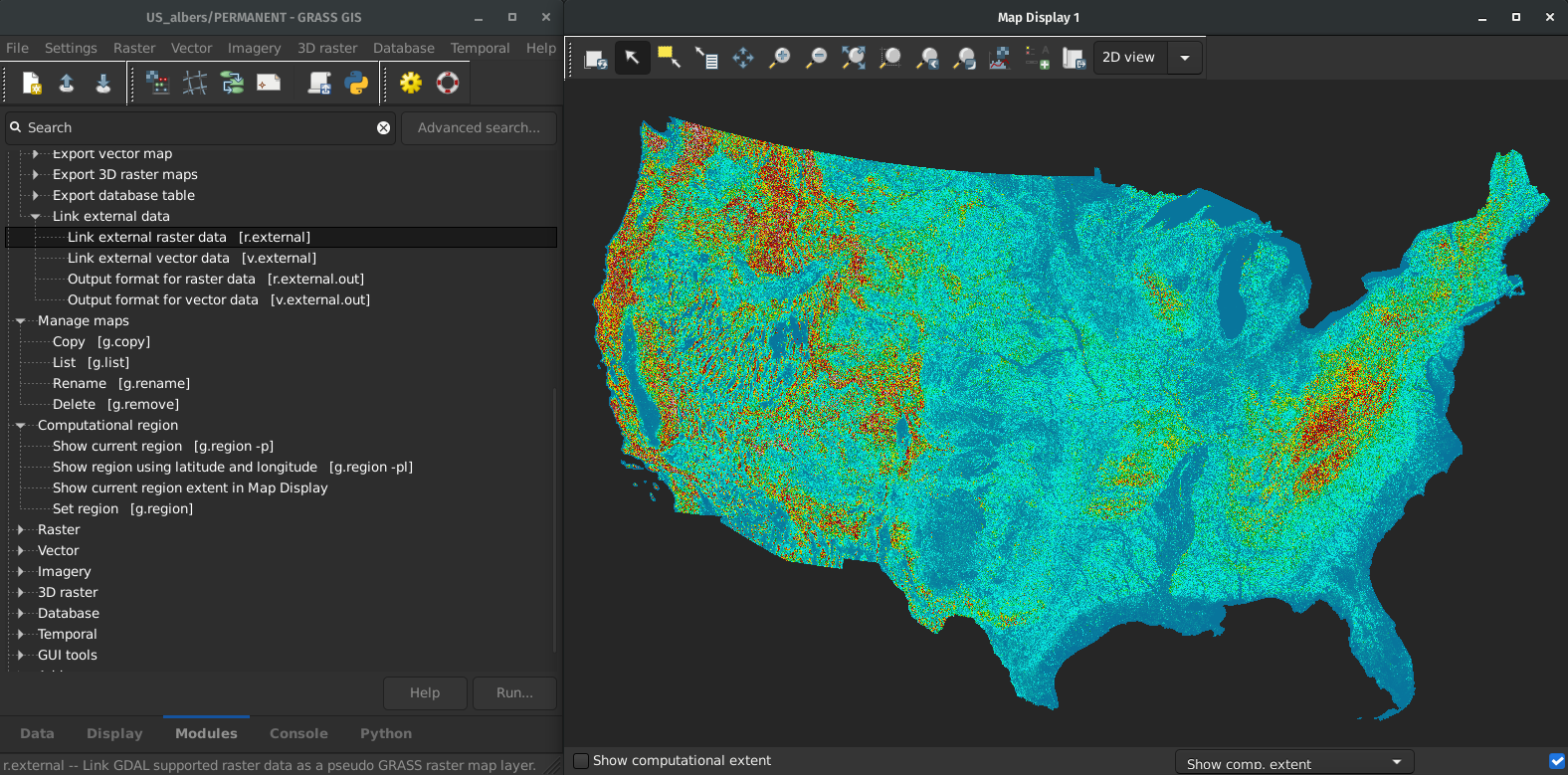
by Markus Metz
Great for workflows when only portion of the data is processed in GRASS GIS.
R packages using GRASS in the background
- hydrographr: wrappers for GDAL and GRASS GIS functions to efficiently work with Hydrography90m and spatial biodiversity data.
- rdwplus: implementation of the IDW-PLUS (inverse distance weighted percent land use for streams) algorithm with GRASS GIS tools.
- fasterRaster: handle large-in-memory/large-on-disk spatial rasters and vectors in R through GRASS GIS.

Only a (dedicated) subset of GRASS tools available through these packages
R packages interfacing with GRASS
- qgisprocess: provides an R interface to the geoprocessing algorithms of QGIS, included GRASS provider (only a selection).
- link2GI: functions to connect to common open source GI software packages, via wrapper functions and direct API-use with system calls.
- rgrass: interpreted interface between GRASS GIS and R, provides facilities for using all GRASS tools from the R command line.
rgrass
initGRASS(): starts a GRASS session from RexecGRASS(): executes GRASS commands from Rread_VECT()andread_RAST(): read vector and raster maps from a GRASS project into Rwrite_VECT()andwrite_RAST(): write vector and raster objects from R into a GRASS project
Access the full stack of tools and extensions in GRASS GIS from R!!
How to use GRASS through the rgrass package?

Choose your own adventure!
Use GRASS GIS tools within your R spatial workflows
- Create a temporary GRASS project with the raster or vector map you want to process with GRASS
- Write the R map object into the GRASS project
- Run the desired GRASS tool(s) from R
- Export the results back to R or your disk
Use GRASS GIS tools within your R spatial workflows
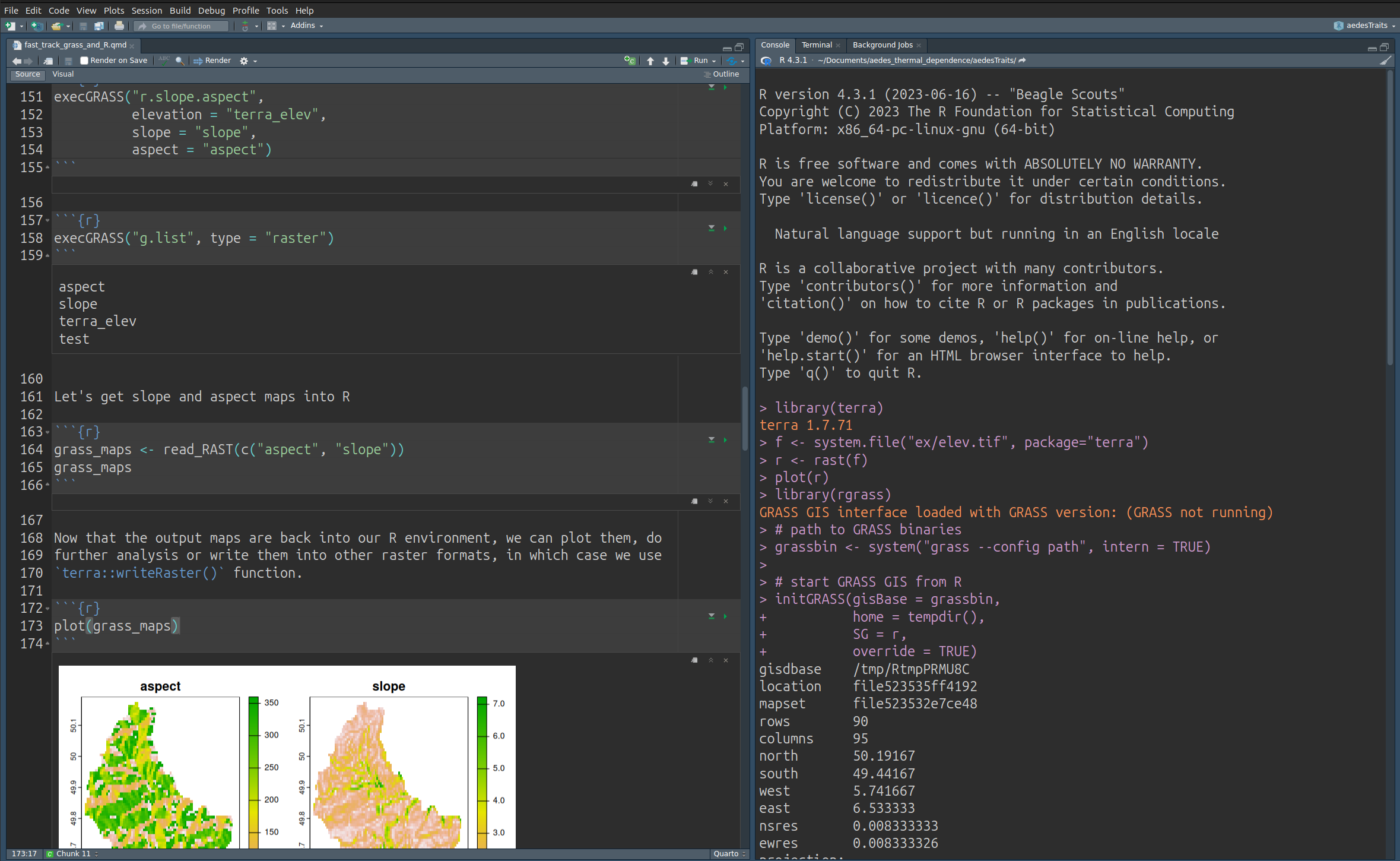
Assuming we have a terra SpatRast object, r, within our R session:
library(rgrass)
grassbin <- system("grass --config path", intern = TRUE)
initGRASS(gisBase = grassbin,
home = tempdir(),
SG = r,
override = TRUE)
write_RAST(r, "terra_elev")
execGRASS("r.slope.aspect",
elevation = "terra_elev",
slope = "slope",
aspect = "aspect")
grass_maps <- read_RAST(c("aspect", "slope"))
Alternatively...
execGRASS("r.import", input=r, output="terra_elev")
execGRASS("r.external", input="file.tif", output="file") # much faster
Use R tools within GRASS GIS workflows
- From R, start GRASS within your project
- Read vector or raster data from GRASS into R
- Do your analysis, modeling, visualizations with R tools
- Optionally write vector and raster outputs back into GRASS
Use R tools within GRASS GIS workflows
grassdata <- path.expand("~/grassdata/")
project <- "nc_basic_spm_grass7"
mapset <- "PERMANENT"
initGRASS(gisBase = grassbin,,
gisDbase = grassdata,
location = project,
mapset = mapset,
override = TRUE,
remove_GISRC= TRUE)
execGRASS("g.list", parameters = list(type="raster"))
elev <- read_RAST("elevation")
# ... plot, run analysis and models, get outputs
write_RAST(elev, "new_elevation")
Support for gdal-grass driver added to read directly from GRASS projects without exporting, thanks to Floris Vanderhaeghe
Use R tools within GRASS GIS workflows: an example
GRASS resources
- Documentation:
- Tutorials (my three favorites):
Support
- Community support
- Commercial support

Take home messages
- Let's (re)use the best tools available for the tasks at hand
- Let's focus on enhancing bridges!


Ask a question, get a special sticker
veroandreo@gmail.com
veroandreo.github.io/grass-gis-talks
Funded by NSF award 2303651